Statistics
http://zzz.bwh.harvard.edu/plink/summary.shtml
Missing genotype
$ plink --bfile mydata --allow-no-sex --missing
# N_MISS (=Number of missing SNPs),
# N_GENO (=Number of non-obligatory missing genotypes),
# F_MISS (=Proportion of missing SNPs).
$ less plink.lmiss
CHR SNP N_MISS N_GENO F_MISS
1 N0 0 1600 0
1 N1 0 1600 0
1 N2 0 1600 0
1 N3 0 1600 0
1 N4 0 1600 0
1 N5 0 1600 0
1 N6 0 1600 0
1 N7 0 1600 0
1 N8 0 1600 0
1 N9 1 1600 0.0113
1 N10 0 1600 0
1 N11 1 1600 0.00565
1 N12 0 1600 0
1 N13 0 1600 0
1 N14 0 1600 0
1 N15 0 1600 0
1 N16 0 1600 0
1 N17 0 1600 0
1 P1 0 1600 0
1 P2 0 1600 0
$ less plink.imiss
FID IID MISS_PHENO N_MISS N_GENO F_MISS
ind_1 ind_1 N 0 20 0
ind_2 ind_2 N 0 20 0
ind_3 ind_3 N 0 20 0
ind_4 ind_4 N 1 20 0.001074
ind_5 ind_5 N 0 20 0
ind_6 ind_6 N 0 20 0
ind_7 ind_7 N 0 20 0
ind_8 ind_8 N 1 20 0.001074
ind_9 ind_9 N 0 20 0
ind_10 ind_10 N 0 20 0
...
$ plink --bfile mydata --allow-no-sex --test-missing
$ less plink.missing
CHR SNP F_MISS_A F_MISS_U P
1 N9 0 0.0113 0.537
1 N11 0 0.00565 1
The way to exclude 10% missing genotype (default):
$ plink --file mydata --mind 0.1 --recode --out cleaned
cleaned.ped # generated
cleaned.map # generated
or
$ plink --file data --mind 0.1 --make-bed --out cleaned
cleaned.bed # generated
cleaned.bim # generated
cleaned.fam # generated
Allele Frequency
- MAF (=Minor Allele Frequency) = P(geno=1, Aa)x0.5 + P(geno=2, aa)
$ plink --bfile mydata --allow-no-sex --freq
$ less plink.frq
# A1 (=Minor allele code), A2 (=Major allele code).
CHR SNP A1 A2 MAF NCHROBS
1 N0 a A 0.01031 3200
1 N1 a A 0.1306 3200
1 N2 a A 0.3644 3200
1 N3 a A 0.4447 3200
1 N4 a A 0.1772 3200
1 N5 a A 0.3956 3200
1 N6 a A 0.2512 3200
1 N7 a A 0.3644 3200
1 N8 a A 0.1734 3200
1 N9 a A 0.3809 3200
1 N10 a A 0.01406 3200
1 N11 a A 0.43 3200
1 N12 a A 0.2391 3200
1 N13 a A 0.03812 3200
1 N14 a A 0.08906 3200
1 N15 a A 0.3331 3200
1 N16 a A 0.2128 3200
1 N17 a A 0.3894 3200
1 P1 a A 0.2031 3200
1 P2 a A 0.2022 3200
Only include SNPs with '0.01 <= MAF <= 0.4' :
$ plink --file mydata --make-bed --maf 0.01 --max-maf 0.4 --out mydata_maf
Hardy-Weinberg Equilibrium
$ plink --bfile mydata --allow-no-sex --hardy
# GENO (=Genotype counts: 11/12/22),
# A1 (=Minor allele code),
# A2 (=Major allele code),
# GENO (Genotype counts 11/12/22),
# O(HET) (=Observed heterozygosity),
# E(HET) (=Expected heterozygosity),
# P (=H-W p-value).
$ less plink.hwe
CHR SNP TEST A1 A2 GENO O(HET) E(HET) P
1 N0 ALL a A 0/33/1567 0.02062 0.02041 1
1 N0 AFF a A 0/15/785 0.01875 0.01857 1
1 N0 UNAFF a A 0/18/782 0.0225 0.02225 1
1 N1 ALL a A 26/366/1208 0.2288 0.2271 0.9122
1 N1 AFF a A 17/191/592 0.2388 0.2417 0.7693
1 N1 UNAFF a A 9/175/616 0.2188 0.2121 0.5037
1 N2 ALL a A 205/756/639 0.4725 0.4632 0.4499
1 N2 AFF a A 98/376/326 0.47 0.4594 0.5388
1 N2 UNAFF a A 107/380/313 0.475 0.4668 0.65
1 N3 ALL a A 316/791/493 0.4944 0.4939 1
1 N3 AFF a A 160/399/241 0.4988 0.4949 0.8864
1 N3 UNAFF a A 156/392/252 0.49 0.4928 0.886
1 N4 ALL a A 36/495/1069 0.3094 0.2916 0.01606
1 N4 AFF a A 13/250/537 0.3125 0.2855 0.006323
1 N4 UNAFF a A 23/245/532 0.3062 0.2976 0.4762
1 N5 ALL a A 255/756/589 0.4725 0.4782 0.6381
1 N5 AFF a A 122/372/306 0.465 0.4736 0.6019
1 N5 UNAFF a A 133/384/283 0.48 0.4824 0.8836
1 N6 ALL a A 103/598/899 0.3738 0.3762 0.7905
1 N6 AFF a A 50/299/451 0.3738 0.3744 1
1 N6 UNAFF a A 53/299/448 0.3738 0.3781 0.779
1 N7 ALL a A 217/732/651 0.4575 0.4632 0.6273
1 N7 AFF a A 106/368/326 0.46 0.4622 0.8787
1 N7 UNAFF a A 111/364/325 0.455 0.4642 0.594
1 N8 ALL a A 52/451/1097 0.2819 0.2867 0.486
1 N8 AFF a A 28/218/554 0.2725 0.2838 0.2621
1 N8 UNAFF a A 24/233/543 0.2912 0.2896 1
1 N9 ALL a A 240/739/621 0.4619 0.4716 0.4265
1 N9 AFF a A 109/365/326 0.4562 0.4632 0.7027
1 N9 UNAFF a A 131/374/295 0.4675 0.479 0.5067
1 N10 ALL a A 0/45/1555 0.02812 0.02773 1
1 N10 AFF a A 0/22/778 0.0275 0.02712 1
1 N10 UNAFF a A 0/23/777 0.02875 0.02834 1
1 N11 ALL a A 310/756/534 0.4725 0.4902 0.1533
1 N11 AFF a A 161/387/252 0.4838 0.4935 0.5673
1 N11 UNAFF a A 149/369/282 0.4612 0.4862 0.1463
1 N12 ALL a A 94/577/929 0.3606 0.3638 0.7312
1 N12 AFF a A 47/293/460 0.3662 0.3667 1
1 N12 UNAFF a A 47/284/469 0.355 0.3609 0.6252
1 N13 ALL a A 2/118/1480 0.07375 0.07334 1
1 N13 AFF a A 2/53/745 0.06625 0.06871 0.2632
1 N13 UNAFF a A 0/65/735 0.08125 0.07795 0.6355
1 N14 ALL a A 10/265/1325 0.1656 0.1623 0.5362
1 N14 AFF a A 5/129/666 0.1612 0.1587 0.8242
1 N14 UNAFF a A 5/136/659 0.17 0.1658 0.6684
1 N15 ALL a A 168/730/702 0.4562 0.4443 0.3111
1 N15 AFF a A 83/370/347 0.4625 0.4456 0.3028
1 N15 UNAFF a A 85/360/355 0.45 0.443 0.6903
1 N16 ALL a A 70/541/989 0.3381 0.335 0.7655
1 N16 AFF a A 39/263/498 0.3288 0.3354 0.5979
1 N16 UNAFF a A 31/278/491 0.3475 0.3347 0.3412
1 N17 ALL a A 258/730/612 0.4562 0.4755 0.1034
1 N17 AFF a A 120/370/310 0.4625 0.4718 0.5999
1 N17 UNAFF a A 138/360/302 0.45 0.479 0.08964
1 P1 ALL a A 75/500/1025 0.3125 0.3237 0.1648
1 P1 AFF a A 42/253/505 0.3162 0.3325 0.1672
1 P1 UNAFF a A 33/247/520 0.3088 0.3147 0.5752
1 P2 ALL a A 60/527/1013 0.3294 0.3226 0.4386
1 P2 AFF a A 29/264/507 0.33 0.3215 0.51
1 P2 UNAFF a A 31/263/506 0.3288 0.3237 0.7433
Option to extract data with P <= 0.1:
$ plink --bfile mydata --hwe 0.1 # extract
$ plink --bfile mydata --hwe2 0.1 # asymptotic
Association
http://zzz.bwh.harvard.edu/plink/anal.shtml
$ plink --bfile mydata --allow-no-sex --assoc --ci 0.95
# A1 (=Minor allele name (based on whole sample)),
# F_A (=Frequency of minor allele in cases),
# F_U (=Frequency of minor allele in controls),
# A2 (=Major allele name),
# basic allelic test chi-square (1df), an asymptotic p-value, estimated OR for A1,
# SE (=Standard error),
# L95 (=Lower bound of Confidential Interval of 95%),
# U95 (=Upper bound of Confidential Interval of 95%).
$ less plink.assoc
CHR SNP BP A1 F_A F_U A2 CHISQ P OR SE L95 U95
1 N0 0 a 0.009375 0.01125 A 0.2756 0.5996 0.8318 0.3514 0.4177 1.656
1 N1 1 a 0.1406 0.1206 A 2.818 0.09322 1.193 0.1052 0.9707 1.466
1 N2 2 a 0.3575 0.3712 A 0.653 0.419 0.9424 0.07347 0.816 1.088
1 N3 3 a 0.4494 0.44 A 0.2847 0.5936 1.039 0.07115 0.9035 1.194
1 N4 4 a 0.1725 0.1819 A 0.4823 0.4874 0.9377 0.09262 0.782 1.124
1 N5 5 a 0.385 0.4062 A 1.511 0.219 0.9149 0.07232 0.794 1.054
1 N6 6 a 0.2494 0.2531 A 0.0598 0.8068 0.9803 0.08152 0.8355 1.15
1 N7 7 a 0.3625 0.3662 A 0.04857 0.8256 0.9839 0.07347 0.852 1.136
1 N8 8 a 0.1712 0.1756 A 0.1068 0.7438 0.9699 0.09338 0.8077 1.165
1 N9 9 a 0.3644 0.3975 A 3.722 0.05369 0.8689 0.07286 0.7533 1.002
1 N10 10 a 0.01375 0.01438 A 0.02254 0.8807 0.9559 0.3003 0.5306 1.722
1 N11 11 a 0.4431 0.4169 A 2.249 0.1337 1.113 0.07144 0.9676 1.28
1 N12 12 a 0.2419 0.2362 A 0.1391 0.7091 1.031 0.0829 0.8767 1.213
1 N13 13 a 0.03562 0.04062 A 0.5454 0.4602 0.8724 0.185 0.6071 1.254
1 N14 14 a 0.08688 0.09125 A 0.1887 0.664 0.9475 0.1242 0.7428 1.209
1 N15 15 a 0.335 0.3312 A 0.05064 0.822 1.017 0.07501 0.878 1.178
1 N16 16 a 0.2131 0.2125 A 0.001865 0.9655 1.004 0.08638 0.8474 1.189
1 N17 17 a 0.3812 0.3975 A 0.8885 0.3459 0.9339 0.07252 0.8102 1.077
1 P1 18 a 0.2106 0.1956 A 1.112 0.2916 1.097 0.08793 0.9235 1.303
1 P2 19 a 0.2012 0.2031 A 0.01744 0.8949 0.9884 0.08803 0.8318 1.175
$ plink --bfile mydata --allow-no-sex --fisher # with an exact p-value.
$ less plink.assoc.fisher
CHR SNP BP A1 F_A F_U A2 P OR
1 N0 0 a 0.009375 0.01125 A 0.727 0.8318
1 N1 1 a 0.1406 0.1206 A 0.1038 1.193
1 N2 2 a 0.3575 0.3712 A 0.4405 0.9424
1 N3 3 a 0.4494 0.44 A 0.6185 1.039
1 N4 4 a 0.1725 0.1819 A 0.5169 0.9377
1 N5 5 a 0.385 0.4062 A 0.2329 0.9149
1 N6 6 a 0.2494 0.2531 A 0.8385 0.9803
1 N7 7 a 0.3625 0.3662 A 0.8543 0.9839
1 N8 8 a 0.1712 0.1756 A 0.7794 0.9699
1 N9 9 a 0.3644 0.3975 A 0.05835 0.8689
1 N10 10 a 0.01375 0.01438 A 1 0.9559
1 N11 11 a 0.4431 0.4169 A 0.1432 1.113
1 N12 12 a 0.2419 0.2362 A 0.7402 1.031
1 N13 13 a 0.03562 0.04062 A 0.5183 0.8724
1 N14 14 a 0.08688 0.09125 A 0.7097 0.9475
1 N15 15 a 0.335 0.3312 A 0.8513 1.017
1 N16 16 a 0.2131 0.2125 A 1 1.004
1 N17 17 a 0.3812 0.3975 A 0.3648 0.9339
1 P1 18 a 0.2106 0.1956 A 0.3122 1.097
1 P2 19 a 0.2012 0.2031 A 0.9299 0.9884
$ plink --bfile mydata --allow-no-sex --model
# with an asymptotic p-value, DF (= degrees of freedom)
$ less plink.model
CHR SNP A1 A2 TEST AFF UNAFF CHISQ DF P
1 N0 a A GENO 0/15/785 0/18/782 NA NA NA
1 N0 a A TREND 15/1585 18/1582 0.2785 1 0.5977
1 N0 a A ALLELIC 15/1585 18/1582 0.2756 1 0.5996
1 N0 a A DOM 15/785 18/782 NA NA NA
1 N0 a A REC 0/800 0/800 NA NA NA
1 N1 a A GENO 17/191/592 9/175/616 3.638 2 0.1622
1 N1 a A TREND 225/1375 193/1407 2.838 1 0.09205
1 N1 a A ALLELIC 225/1375 193/1407 2.818 1 0.09322
1 N1 a A DOM 208/592 184/616 1.946 1 0.163
1 N1 a A REC 17/783 9/791 2.502 1 0.1137
1 N2 a A GENO 98/376/326 107/380/313 0.6808 2 0.7115
1 N2 a A TREND 572/1028 594/1006 0.6664 1 0.4143
1 N2 a A ALLELIC 572/1028 594/1006 0.653 1 0.419
1 N2 a A DOM 474/326 487/313 0.4403 1 0.507
1 N2 a A REC 98/702 107/693 0.4532 1 0.5008
1 N3 a A GENO 160/399/241 156/392/252 0.358 2 0.8361
1 N3 a A TREND 719/881 704/896 0.285 1 0.5934
1 N3 a A ALLELIC 719/881 704/896 0.2847 1 0.5936
1 N3 a A DOM 559/241 548/252 0.3547 1 0.5514
1 N3 a A REC 160/640 156/644 0.06309 1 0.8017
1 N4 a A GENO 13/250/537 23/245/532 2.852 2 0.2403
1 N4 a A TREND 276/1324 291/1309 0.5136 1 0.4736
1 N4 a A ALLELIC 276/1324 291/1309 0.4823 1 0.4874
1 N4 a A DOM 263/537 268/532 0.07047 1 0.7907
1 N4 a A REC 13/787 23/777 2.842 1 0.09185
1 N5 a A GENO 122/372/306 133/384/283 1.563 2 0.4577
1 N5 a A TREND 616/984 650/950 1.493 1 0.2218
1 N5 a A ALLELIC 616/984 650/950 1.511 1 0.219
1 N5 a A DOM 494/306 517/283 1.421 1 0.2332
1 N5 a A REC 122/678 133/667 0.5645 1 0.4525
1 N6 a A GENO 50/299/451 53/299/448 0.09739 2 0.9525
1 N6 a A TREND 399/1201 405/1195 0.05941 1 0.8074
1 N6 a A ALLELIC 399/1201 405/1195 0.0598 1 0.8068
1 N6 a A DOM 349/451 352/448 0.02285 1 0.8798
1 N6 a A REC 50/750 53/747 0.09339 1 0.7599
1 N7 a A GENO 106/368/326 111/364/325 0.1386 2 0.933
1 N7 a A TREND 580/1020 586/1014 0.04798 1 0.8266
1 N7 a A ALLELIC 580/1020 586/1014 0.04857 1 0.8256
1 N7 a A DOM 474/326 475/325 0.00259 1 0.9594
1 N7 a A REC 106/694 111/689 0.1333 1 0.7151
1 N8 a A GENO 28/218/554 24/233/543 0.9169 2 0.6323
1 N8 a A TREND 274/1326 281/1319 0.105 1 0.7459
1 N8 a A ALLELIC 274/1326 281/1319 0.1068 1 0.7438
1 N8 a A DOM 246/554 257/543 0.3509 1 0.5536
1 N8 a A REC 28/772 24/776 0.318 1 0.5728
1 N9 a A GENO 109/365/326 131/374/295 3.674 2 0.1593
1 N9 a A TREND 583/1017 636/964 3.647 1 0.05618
1 N9 a A ALLELIC 583/1017 636/964 3.722 1 0.05369
1 N9 a A DOM 474/326 505/295 2.529 1 0.1118
1 N9 a A REC 109/691 131/669 2.373 1 0.1235
1 N10 a A GENO 0/22/778 0/23/777 NA NA NA
1 N10 a A TREND 22/1578 23/1577 0.02287 1 0.8798
1 N10 a A ALLELIC 22/1578 23/1577 0.02254 1 0.8807
1 N10 a A DOM 22/778 23/777 NA NA NA
1 N10 a A REC 0/800 0/800 NA NA NA
1 N11 a A GENO 161/387/252 149/369/282 2.578 2 0.2755
1 N11 a A TREND 709/891 667/933 2.171 1 0.1407
1 N11 a A ALLELIC 709/891 667/933 2.249 1 0.1337
1 N11 a A DOM 548/252 518/282 2.53 1 0.1117
1 N11 a A REC 161/639 149/651 0.5761 1 0.4478
1 N12 a A GENO 47/293/460 47/284/469 0.2276 2 0.8924
1 N12 a A TREND 387/1213 378/1222 0.1379 1 0.7103
1 N12 a A ALLELIC 387/1213 378/1222 0.1391 1 0.7091
1 N12 a A DOM 340/460 331/469 0.2079 1 0.6484
1 N12 a A REC 47/753 47/753 0 1 1
1 N13 a A GENO 2/53/745 0/65/735 NA NA NA
1 N13 a A TREND 57/1543 65/1535 0.5484 1 0.459
1 N13 a A ALLELIC 57/1543 65/1535 0.5454 1 0.4602
1 N13 a A DOM 55/745 65/735 NA NA NA
1 N13 a A REC 2/798 0/800 NA NA NA
1 N14 a A GENO 5/129/666 5/136/659 0.2219 2 0.895
1 N14 a A TREND 139/1461 146/1454 0.1927 1 0.6606
1 N14 a A ALLELIC 139/1461 146/1454 0.1887 1 0.664
1 N14 a A DOM 134/666 141/659 0.2152 1 0.6428
1 N14 a A REC 5/795 5/795 0 1 1
1 N15 a A GENO 83/370/347 85/360/355 0.252 2 0.8816
1 N15 a A TREND 536/1064 530/1070 0.05204 1 0.8196
1 N15 a A ALLELIC 536/1064 530/1070 0.05064 1 0.822
1 N15 a A DOM 453/347 445/355 0.1624 1 0.6869
1 N15 a A REC 83/717 85/715 0.0266 1 0.8704
1 N16 a A GENO 39/263/498 31/278/491 1.38 2 0.5016
1 N16 a A TREND 341/1259 340/1260 0.001883 1 0.9654
1 N16 a A ALLELIC 341/1259 340/1260 0.001865 1 0.9655
1 N16 a A DOM 302/498 309/491 0.1297 1 0.7187
1 N16 a A REC 39/761 31/769 0.9561 1 0.3282
1 N17 a A GENO 120/370/310 138/360/302 1.497 2 0.473
1 N17 a A TREND 610/990 636/964 0.8539 1 0.3555
1 N17 a A ALLELIC 610/990 636/964 0.8885 1 0.3459
1 N17 a A DOM 490/310 498/302 0.1694 1 0.6807
1 N17 a A REC 120/680 138/662 1.497 1 0.2211
1 P1 a A GENO 42/253/505 33/247/520 1.372 2 0.5037
1 P1 a A TREND 337/1263 313/1287 1.075 1 0.2999
1 P1 a A ALLELIC 337/1263 313/1287 1.112 1 0.2916
1 P1 a A DOM 295/505 280/520 0.6108 1 0.4345
1 P1 a A REC 42/758 33/767 1.133 1 0.2871
1 P2 a A GENO 29/264/507 31/263/506 0.06955 2 0.9658
1 P2 a A TREND 322/1278 325/1275 0.01781 1 0.8938
1 P2 a A ALLELIC 322/1278 325/1275 0.01744 1 0.8949
1 P2 a A DOM 293/507 294/506 0.002691 1 0.9586
1 P2 a A REC 29/771 31/769 0.06926 1 0.7924
$ plink --bfile mydata --allow-no-sex --assoc --adjust qq-plot
# UNADJ (=Unadjusted p-value),
# GC (=Genomic-control corrected p-values),
# BONF (=Bonferroni single-step adjusted p-values),
# HOLM (=Holm (1979) step-down adjusted p-values),
# SIDAK_SS (=Sidak single-step adjusted p-values),
# SIDAK_SD (=Sidak step-down adjusted p-values),
# FDR_BH (=Benjamini & Hochberg (1995) step-up FDR control),
# FDR_BY (=Benjamini & Yekutieli (2001) step-up FDR control).
$ less plink.assoc.adjusted
CHR SNP UNADJ GC QQ BONF HOLM SIDAK_SS SIDAK_SD FDR_BH FDR_BY
1 N9 0.05369 0.05369 0.025 1 1 0.6684 0.6684 0.8913 1
1 N1 0.09322 0.09322 0.075 1 1 0.8587 0.8442 0.8913 1
1 N11 0.1337 0.1337 0.125 1 1 0.9433 0.9245 0.8913 1
1 N5 0.219 0.219 0.175 1 1 0.9929 0.985 0.9421 1
1 P1 0.2916 0.2916 0.225 1 1 0.999 0.996 0.9421 1
1 N17 0.3459 0.3459 0.275 1 1 0.9998 0.9983 0.9421 1
1 N2 0.419 0.419 0.325 1 1 1 0.9995 0.9421 1
1 N13 0.4602 0.4602 0.375 1 1 1 0.9997 0.9421 1
1 N4 0.4874 0.4874 0.425 1 1 1 0.9997 0.9421 1
1 N3 0.5936 0.5936 0.475 1 1 1 1 0.9421 1
1 N0 0.5996 0.5996 0.525 1 1 1 1 0.9421 1
1 N14 0.664 0.664 0.575 1 1 1 1 0.9421 1
1 N12 0.7091 0.7091 0.625 1 1 1 1 0.9421 1
1 N8 0.7438 0.7438 0.675 1 1 1 1 0.9421 1
1 N6 0.8068 0.8068 0.725 1 1 1 1 0.9421 1
1 N15 0.822 0.822 0.775 1 1 1 1 0.9421 1
1 N7 0.8256 0.8256 0.825 1 1 1 1 0.9421 1
1 N10 0.8807 0.8807 0.875 1 1 1 1 0.9421 1
1 P2 0.8949 0.8949 0.925 1 1 1 1 0.9421 1
1 N16 0.9655 0.9655 0.975 1 1 1 1 0.9655 1
QQ-plot in R
> data<-read.table(file="plink.assoc.adjusted", header=T)
> plot(-log(data$QQ, 10), -log(data$UNADJ,10), xlab ="expected –logP values", ylab = "observed –logP values")
> abline(a=0, b=1)
$ plink --bfile mydata --allow-no-sex -logistic beta
# with NMISS (=Number of non-missing individuals included in analysis),
# BETA (=Regression coefficient (--linear) or odds ratio (--logistic)),
# STAT (=Coefficient t-statistics),
# an asymptotic p-value for t-statistics.
$ less plink.assoc.logistic
CHR SNP BP A1 TEST NMISS BETA STAT P
1 N0 0 a ADD 1600 -0.1861 -0.527 0.5982
1 N1 1 a ADD 1600 0.1778 1.683 0.09247
1 N2 2 a ADD 1600 -0.0606 -0.8162 0.4144
1 N3 3 a ADD 1600 0.03801 0.5339 0.5934
1 N4 4 a ADD 1600 -0.06851 -0.7166 0.4736
1 N5 5 a ADD 1600 -0.08788 -1.221 0.2219
1 N6 6 a ADD 1600 -0.0198 -0.2438 0.8074
1 N7 7 a ADD 1600 -0.01599 -0.219 0.8266
1 N8 8 a ADD 1600 -0.03002 -0.3241 0.7459
1 N9 9 a ADD 1600 -0.1378 -1.908 0.05637
1 N10 10 a ADD 1600 -0.04574 -0.1512 0.8798
1 N11 11 a ADD 1600 0.1035 1.473 0.1408
1 N12 12 a ADD 1600 0.03066 0.3714 0.7103
1 N13 13 a ADD 1600 -0.1373 -0.74 0.4593
1 N14 14 a ADD 1600 -0.05508 -0.439 0.6607
1 N15 15 a ADD 1600 0.01735 0.2282 0.8195
1 N16 16 a ADD 1600 0.00376 0.04333 0.9654
1 N17 17 a ADD 1600 -0.0657 -0.9239 0.3556
1 P1 18 a ADD 1600 0.08963 1.036 0.3
1 P2 19 a ADD 1600 -0.01188 -0.1335 0.8938
LD calculation
D': If D'=0.8, it means that basically the two SNPs are coinherited roughly 80% of the time. https://www.researchgate.net/post/I-have-Linkage-Disequilibrium-LD-data-for-two-SNPs-r2-is-about-014-D-is-around-08-Could-these-SNPs-be-said-to-be-in-strong-LD
http://zzz.bwh.harvard.edu/plink/ld.shtml
$ plink --bfile mydata --allow-no-sex --r2 dprime inter-chr with-freqs --ld-window-r2 0.005
$ less plink.ld
CHR_A BP_A SNP_A MAF_A CHR_B BP_B SNP_B MAF_B R2 DP
1 5 N5 0.395625 1 19 P2 0.202188 0.00611992 0.125729
1 18 P1 0.203125 1 19 P2 0.202188 0.0279074 0.16754
$ plink --bfile mydata --allow-no-sex --r2 d inter-chr with-freqs --ld-window-r2 0.005
$ less plink.ld
CHR_A BP_A SNP_A MAF_A CHR_B BP_B SNP_B MAF_B R2 D
1 5 N5 0.395625 1 19 P2 0.202188 0.00611992 0.0153637
1 18 P1 0.203125 1 19 P2 0.202188 0.0279074 0.0269938
$ plink --bfile mydata --allow-no-sex --r2 dprime inter-chr with-freqs --ld-window-r2 0.001
$ less plink.ld
CHR_A BP_A SNP_A MAF_A CHR_B BP_B SNP_B MAF_B R2 DP
1 0 N0 0.0103125 1 8 N8 0.173438 0.00173647 0.186997
1 0 N0 0.0103125 1 9 N9 0.380938 0.00107551 0.409557
1 0 N0 0.0103125 1 14 N14 0.0890625 0.00101876 1
1 0 N0 0.0103125 1 16 N16 0.212812 0.00312607 0.284791
1 0 N0 0.0103125 1 19 P2 0.202188 0.00209782 0.891301
1 1 N1 0.130625 1 2 N2 0.364375 0.0017376 0.142034
1 1 N1 0.130625 1 3 N3 0.444688 0.00216632 0.134182
1 1 N1 0.130625 1 4 N4 0.177188 0.00119479 0.192163
1 1 N1 0.130625 1 12 N12 0.239062 0.00138925 0.0538967
1 1 N1 0.130625 1 14 N14 0.0890625 0.00265773 0.063909
1 1 N1 0.130625 1 17 N17 0.389375 0.00150455 0.079908
1 2 N2 0.364375 1 6 N6 0.25125 0.00159545 0.0522072
1 2 N2 0.364375 1 11 N11 0.43 0.00327338 0.0870017
1 2 N2 0.364375 1 17 N17 0.389375 0.00105073 0.0341876
1 2 N2 0.364375 1 19 P2 0.202188 0.00104851 0.0487004
1 3 N3 0.444688 1 8 N8 0.173438 0.0015051 0.0757893
1 3 N3 0.444688 1 10 N10 0.0140625 0.00213762 0.432614
1 3 N3 0.444688 1 16 N16 0.212812 0.00136734 0.0794732
1 3 N3 0.444688 1 19 P2 0.202188 0.00175142 0.0743921
1 4 N4 0.177188 1 6 N6 0.25125 0.00120982 0.129392
1 4 N4 0.177188 1 9 N9 0.380938 0.00134834 0.100873
1 4 N4 0.177188 1 12 N12 0.239062 0.00206632 0.174764
1 4 N4 0.177188 1 14 N14 0.0890625 0.00104996 0.0480894
1 4 N4 0.177188 1 17 N17 0.389375 0.00364646 0.162957
1 5 N5 0.395625 1 9 N9 0.380938 0.00151254 0.0612781
1 5 N5 0.395625 1 12 N12 0.239062 0.00159792 0.0577013
1 5 N5 0.395625 1 15 N15 0.333125 0.00149228 0.0442214
1 5 N5 0.395625 1 16 N16 0.212812 0.0015976 0.062196
1 5 N5 0.395625 1 19 P2 0.202188 0.00611992 0.125729
1 7 N7 0.364375 1 8 N8 0.173438 0.00269075 0.149565
1 7 N7 0.364375 1 11 N11 0.43 0.00107766 0.0376586
1 7 N7 0.364375 1 17 N17 0.389375 0.00159406 0.042109
1 8 N8 0.173438 1 17 N17 0.389375 0.00132136 0.0993757
1 9 N9 0.380938 1 13 N13 0.038125 0.00382624 0.243725
1 10 N10 0.0140625 1 17 N17 0.389375 0.0023212 0.505188
1 10 N10 0.0140625 1 18 P1 0.203125 0.00101458 0.134655
1 11 N11 0.43 1 15 N15 0.333125 0.0019084 0.0536846
1 11 N11 0.43 1 17 N17 0.389375 0.00196082 0.0638448
1 13 N13 0.038125 1 16 N16 0.212812 0.00103394 0.0839774
1 14 N14 0.0890625 1 16 N16 0.212812 0.00109154 0.203216
1 14 N14 0.0890625 1 17 N17 0.389375 0.00135083 0.0938628
1 15 N15 0.333125 1 18 P1 0.203125 0.00209955 0.0641442
1 16 N16 0.212812 1 17 N17 0.389375 0.00168437 0.0630312
1 16 N16 0.212812 1 19 P2 0.202188 0.00195047 0.0456145
1 17 N17 0.389375 1 19 P2 0.202188 0.00103718 0.0801131
1 18 P1 0.203125 1 19 P2 0.202188 0.0279074 0.16754
$ plink --bfile mydata --allow-no-sex --r2 dprime inter-chr with-freqs --ld-window-r2 0
$ less plink.ld
CHR_A BP_A SNP_A MAF_A CHR_B BP_B SNP_B MAF_B R2 DP
1 0 N0 0.0103125 1 1 N1 0.130625 0.000464463 0.0818375
1 0 N0 0.0103125 1 2 N2 0.364375 8.99385e-05 0.122706
1 0 N0 0.0103125 1 3 N3 0.444688 0.000447708 0.231636
1 0 N0 0.0103125 1 4 N4 0.177188 0.000802884 0.598174
1 0 N0 0.0103125 1 5 N5 0.395625 0.000299645 0.209596
1 0 N0 0.0103125 1 6 N6 0.25125 0.000278992 0.0947867
1 0 N0 0.0103125 1 7 N7 0.364375 0.000129386 0.0843693
1 0 N0 0.0103125 1 8 N8 0.173438 0.00173647 0.186997
1 0 N0 0.0103125 1 9 N9 0.380938 0.00107551 0.409557
1 0 N0 0.0103125 1 10 N10 0.0140625 0.000148621 1
1 0 N0 0.0103125 1 11 N11 0.43 7.23778e-06 0.0228911
1 0 N0 0.0103125 1 12 N12 0.239062 0.000665512 0.141653
1 0 N0 0.0103125 1 13 N13 0.038125 0.000636386 0.0492009
1 0 N0 0.0103125 1 14 N14 0.0890625 0.00101876 1
1 0 N0 0.0103125 1 15 N15 0.333125 0.000432297 0.288189
1 0 N0 0.0103125 1 16 N16 0.212812 0.00312607 0.284791
1 0 N0 0.0103125 1 17 N17 0.389375 0.000770346 0.217123
1 0 N0 0.0103125 1 18 P1 0.203125 0.000798405 0.139754
1 0 N0 0.0103125 1 19 P2 0.202188 0.00209782 0.891301
1 1 N1 0.130625 1 2 N2 0.364375 0.0017376 0.142034
1 1 N1 0.130625 1 3 N3 0.444688 0.00216632 0.134182
1 1 N1 0.130625 1 4 N4 0.177188 0.00119479 0.192163
1 1 N1 0.130625 1 5 N5 0.395625 0.000290652 0.0355849
1 1 N1 0.130625 1 6 N6 0.25125 0.000894341 0.133186
1 1 N1 0.130625 1 7 N7 0.364375 0.000412185 0.0396561
1 1 N1 0.130625 1 8 N8 0.173438 0.000414408 0.114649
1 1 N1 0.130625 1 9 N9 0.380938 0.000601054 0.0806283
1 1 N1 0.130625 1 10 N10 0.0140625 0.000327185 0.0587083
1 1 N1 0.130625 1 11 N11 0.43 5.02405e-05 0.0158823
1 1 N1 0.130625 1 12 N12 0.239062 0.00138925 0.0538967
1 1 N1 0.130625 1 13 N13 0.038125 0.000324482 0.0350719
1 1 N1 0.130625 1 14 N14 0.0890625 0.00265773 0.063909
1 1 N1 0.130625 1 15 N15 0.333125 0.000879857 0.108272
1 1 N1 0.130625 1 16 N16 0.212812 0.000476227 0.0292723
1 1 N1 0.130625 1 17 N17 0.389375 0.00150455 0.079908
1 1 N1 0.130625 1 18 P1 0.203125 0.000457656 0.0278642
1 1 N1 0.130625 1 19 P2 0.202188 0.000359164 0.0971203
1 2 N2 0.364375 1 3 N3 0.444688 2.98863e-05 0.0080687
1 2 N2 0.364375 1 4 N4 0.177188 3.69576e-05 0.0173026
1 2 N2 0.364375 1 5 N5 0.395625 0.000200431 0.0231111
1 2 N2 0.364375 1 6 N6 0.25125 0.00159545 0.0522072
1 2 N2 0.364375 1 7 N7 0.364375 5.82913e-06 0.00421167
1 2 N2 0.364375 1 8 N8 0.173438 0.000172933 0.0379167
1 2 N2 0.364375 1 9 N9 0.380938 0.000130041 0.0118148
1 2 N2 0.364375 1 10 N10 0.0140625 0.000399602 0.12673
1 2 N2 0.364375 1 11 N11 0.43 0.00327338 0.0870017
1 2 N2 0.364375 1 12 N12 0.239062 4.76657e-05 0.00932601
1 2 N2 0.364375 1 13 N13 0.038125 8.11514e-05 0.0342591
1 2 N2 0.364375 1 14 N14 0.0890625 1.14382e-05 0.00818936
1 2 N2 0.364375 1 15 N15 0.333125 2.98329e-06 0.00185029
1 2 N2 0.364375 1 16 N16 0.212812 0.000106566 0.0262227
1 2 N2 0.364375 1 17 N17 0.389375 0.00105073 0.0341876
1 2 N2 0.364375 1 18 P1 0.203125 0.000938773 0.0459481
1 2 N2 0.364375 1 19 P2 0.202188 0.00104851 0.0487004
1 3 N3 0.444688 1 4 N4 0.177188 0.000167608 0.0311761
1 3 N3 0.444688 1 5 N5 0.395625 0.000127855 0.0125063
1 3 N3 0.444688 1 6 N6 0.25125 1.75507e-06 0.00204655
1 3 N3 0.444688 1 7 N7 0.364375 2.64163e-05 0.00607465
1 3 N3 0.444688 1 8 N8 0.173438 0.0015051 0.0757893
1 3 N3 0.444688 1 9 N9 0.380938 0.000124643 0.012736
1 3 N3 0.444688 1 10 N10 0.0140625 0.00213762 0.432614
1 3 N3 0.444688 1 11 N11 0.43 7.66189e-05 0.00901841
1 3 N3 0.444688 1 12 N12 0.239062 8.51752e-05 0.0184
1 3 N3 0.444688 1 13 N13 0.038125 0.000781858 0.125683
1 3 N3 0.444688 1 14 N14 0.0890625 1.16081e-05 0.0121764
1 3 N3 0.444688 1 15 N15 0.333125 1.75381e-05 0.00530236
1 3 N3 0.444688 1 16 N16 0.212812 0.00136734 0.0794732
1 3 N3 0.444688 1 17 N17 0.389375 7.15784e-06 0.003744
1 3 N3 0.444688 1 18 P1 0.203125 0.000108528 0.0184647
1 3 N3 0.444688 1 19 P2 0.202188 0.00175142 0.0743921
1 4 N4 0.177188 1 5 N5 0.395625 0.000145965 0.0321788
1 4 N4 0.177188 1 6 N6 0.25125 0.00120982 0.129392
1 4 N4 0.177188 1 7 N7 0.364375 1.01445e-05 0.00519665
1 4 N4 0.177188 1 8 N8 0.173438 8.63245e-05 0.0437086
1 4 N4 0.177188 1 9 N9 0.380938 0.00134834 0.100873
1 4 N4 0.177188 1 10 N10 0.0140625 4.86309e-05 0.12583
1 4 N4 0.177188 1 11 N11 0.43 0.000159189 0.0236149
1 4 N4 0.177188 1 12 N12 0.239062 0.00206632 0.174764
1 4 N4 0.177188 1 13 N13 0.038125 0.000156984 0.0292044
1 4 N4 0.177188 1 14 N14 0.0890625 0.00104996 0.0480894
1 4 N4 0.177188 1 15 N15 0.333125 2.02081e-06 0.00433426
1 4 N4 0.177188 1 16 N16 0.212812 0.000317612 0.0199683
1 4 N4 0.177188 1 17 N17 0.389375 0.00364646 0.162957
1 4 N4 0.177188 1 18 P1 0.203125 8.39521e-06 0.012367
1 4 N4 0.177188 1 19 P2 0.202188 0.000249608 0.0171392
1 5 N5 0.395625 1 6 N6 0.25125 0.00068467 0.0365464
1 5 N5 0.395625 1 7 N7 0.364375 0.000100069 0.01633
1 5 N5 0.395625 1 8 N8 0.173438 0.000620234 0.0439879
1 5 N5 0.395625 1 9 N9 0.380938 0.00151254 0.0612781
1 5 N5 0.395625 1 10 N10 0.0140625 6.98189e-05 0.0864752
1 5 N5 0.395625 1 11 N11 0.43 4.84599e-06 0.00313261
1 5 N5 0.395625 1 12 N12 0.239062 0.00159792 0.0577013
1 5 N5 0.395625 1 13 N13 0.038125 2.33295e-06 0.0062072
1 5 N5 0.395625 1 14 N14 0.0890625 0.000653473 0.0661453
1 5 N5 0.395625 1 15 N15 0.333125 0.00149228 0.0442214
1 5 N5 0.395625 1 16 N16 0.212812 0.0015976 0.062196
1 5 N5 0.395625 1 17 N17 0.389375 0.000751801 0.0277807
1 5 N5 0.395625 1 18 P1 0.203125 0.000258788 0.0393819
1 5 N5 0.395625 1 19 P2 0.202188 0.00611992 0.125729
1 6 N6 0.25125 1 7 N7 0.364375 0.000790317 0.0367443
1 6 N6 0.25125 1 8 N8 0.173438 0.000328831 0.0229318
1 6 N6 0.25125 1 9 N9 0.380938 0.000354788 0.025507
1 6 N6 0.25125 1 10 N10 0.0140625 2.34578e-05 0.0700088
1 6 N6 0.25125 1 11 N11 0.43 0.00091058 0.0452451
1 6 N6 0.25125 1 12 N12 0.239062 0.000128693 0.0117241
1 6 N6 0.25125 1 13 N13 0.038125 0.000171897 0.0381481
1 6 N6 0.25125 1 14 N14 0.0890625 2.78651e-05 0.0291436
1 6 N6 0.25125 1 15 N15 0.333125 2.36397e-05 0.00593223
1 6 N6 0.25125 1 16 N16 0.212812 0.000985341 0.0349718
1 6 N6 0.25125 1 17 N17 0.389375 3.40884e-05 0.0126218
1 6 N6 0.25125 1 18 P1 0.203125 0.000179483 0.0458079
1 6 N6 0.25125 1 19 P2 0.202188 0.000647251 0.0872419
1 7 N7 0.364375 1 8 N8 0.173438 0.00269075 0.149565
1 7 N7 0.364375 1 9 N9 0.380938 0.000601996 0.0254204
1 7 N7 0.364375 1 10 N10 0.0140625 1.8525e-05 0.0475991
1 7 N7 0.364375 1 11 N11 0.43 0.00107766 0.0376586
1 7 N7 0.364375 1 12 N12 0.239062 5.01497e-07 0.0016687
1 7 N7 0.364375 1 13 N13 0.038125 1.44337e-05 0.0144483
1 7 N7 0.364375 1 14 N14 0.0890625 0.000269272 0.0693137
1 7 N7 0.364375 1 15 N15 0.333125 0.000744286 0.0509818
1 7 N7 0.364375 1 16 N16 0.212812 0.000168939 0.0189269
1 7 N7 0.364375 1 17 N17 0.389375 0.00159406 0.042109
1 7 N7 0.364375 1 18 P1 0.203125 0.000130385 0.0171238
1 7 N7 0.364375 1 19 P2 0.202188 0.000339879 0.0277274
1 8 N8 0.173438 1 9 N9 0.380938 0.00081243 0.0488111
1 8 N8 0.173438 1 10 N10 0.0140625 0.000804644 0.1088
1 8 N8 0.173438 1 11 N11 0.43 6.99992e-06 0.0050166
1 8 N8 0.173438 1 12 N12 0.239062 0.000699341 0.102998
1 8 N8 0.173438 1 13 N13 0.038125 9.11273e-06 0.00694564
1 8 N8 0.173438 1 14 N14 0.0890625 0.000746756 0.0400332
1 8 N8 0.173438 1 15 N15 0.333125 4.0685e-05 0.0197016
1 8 N8 0.173438 1 16 N16 0.212812 0.000988257 0.13199
1 8 N8 0.173438 1 17 N17 0.389375 0.00132136 0.0993757
1 8 N8 0.173438 1 18 P1 0.203125 0.000393534 0.0218647
1 8 N8 0.173438 1 19 P2 0.202188 1.09249e-05 0.00363246
1 9 N9 0.380938 1 10 N10 0.0140625 0.000209563 0.0950845
1 9 N9 0.380938 1 11 N11 0.43 4.64625e-05 0.0100045
1 9 N9 0.380938 1 12 N12 0.239062 1.49149e-06 0.0027776
1 9 N9 0.380938 1 13 N13 0.038125 0.00382624 0.243725
1 9 N9 0.380938 1 14 N14 0.0890625 0.000205038 0.0583787
1 9 N9 0.380938 1 15 N15 0.333125 0.000178272 0.014819
1 9 N9 0.380938 1 16 N16 0.212812 5.59517e-05 0.0183395
1 9 N9 0.380938 1 17 N17 0.389375 0.000387392 0.0314209
1 9 N9 0.380938 1 18 P1 0.203125 0.000154354 0.0193033
1 9 N9 0.380938 1 19 P2 0.202188 0.000134158 0.0180485
1 10 N10 0.0140625 1 11 N11 0.43 0.0002223 0.143736
1 10 N10 0.0140625 1 12 N12 0.239062 8.63261e-05 0.0436059
1 10 N10 0.0140625 1 13 N13 0.038125 0.000528059 0.0383072
1 10 N10 0.0140625 1 14 N14 0.0890625 0.000980866 0.0819977
1 10 N10 0.0140625 1 15 N15 0.333125 3.64147e-05 0.0714909
1 10 N10 0.0140625 1 16 N16 0.212812 2.56131e-05 0.0220335
1 10 N10 0.0140625 1 17 N17 0.389375 0.0023212 0.505188
1 10 N10 0.0140625 1 18 P1 0.203125 0.00101458 0.134655
1 10 N10 0.0140625 1 19 P2 0.202188 1.26688e-06 0.0187212
1 11 N11 0.43 1 12 N12 0.239062 0.000161567 0.0196966
1 11 N11 0.43 1 13 N13 0.038125 6.87411e-05 0.0479475
1 11 N11 0.43 1 14 N14 0.0890625 0.000872452 0.10876
1 11 N11 0.43 1 15 N15 0.333125 0.0019084 0.0536846
1 11 N11 0.43 1 16 N16 0.212812 0.000765209 0.0612539
1 11 N11 0.43 1 17 N17 0.389375 0.00196082 0.0638448
1 11 N11 0.43 1 18 P1 0.203125 6.08111e-05 0.0177831
1 11 N11 0.43 1 19 P2 0.202188 0.000263488 0.0371242
1 12 N12 0.239062 1 13 N13 0.038125 0.000295513 0.15405
1 12 N12 0.239062 1 14 N14 0.0890625 0.000249644 0.0901521
1 12 N12 0.239062 1 15 N15 0.333125 0.000399751 0.0252113
1 12 N12 0.239062 1 16 N16 0.212812 0.000508144 0.0243005
1 12 N12 0.239062 1 17 N17 0.389375 0.000632754 0.0562004
1 12 N12 0.239062 1 18 P1 0.203125 0.000139554 0.0417448
1 12 N12 0.239062 1 19 P2 0.202188 1.12005e-05 0.00372626
1 13 N13 0.038125 1 14 N14 0.0890625 0.000784602 0.0439929
1 13 N13 0.038125 1 15 N15 0.333125 0.000106354 0.0732908
1 13 N13 0.038125 1 16 N16 0.212812 0.00103394 0.0839774
1 13 N13 0.038125 1 17 N17 0.389375 9.0432e-05 0.0598162
1 13 N13 0.038125 1 18 P1 0.203125 0.000920573 0.0769432
1 13 N13 0.038125 1 19 P2 0.202188 0.000705199 0.0671486
1 14 N14 0.0890625 1 15 N15 0.333125 7.45231e-06 0.00617055
1 14 N14 0.0890625 1 16 N16 0.212812 0.00109154 0.203216
1 14 N14 0.0890625 1 17 N17 0.389375 0.00135083 0.0938628
1 14 N14 0.0890625 1 18 P1 0.203125 0.000924271 0.0490889
1 14 N14 0.0890625 1 19 P2 0.202188 6.67493e-05 0.0131537
1 15 N15 0.333125 1 16 N16 0.212812 0.000105878 0.013987
1 15 N15 0.333125 1 17 N17 0.389375 2.28392e-06 0.0026777
1 15 N15 0.333125 1 18 P1 0.203125 0.00209955 0.0641442
1 15 N15 0.333125 1 19 P2 0.202188 0.000152973 0.0173645
1 16 N16 0.212812 1 17 N17 0.389375 0.00168437 0.0630312
1 16 N16 0.212812 1 18 P1 0.203125 0.000671612 0.0987218
1 16 N16 0.212812 1 19 P2 0.202188 0.00195047 0.0456145
1 17 N17 0.389375 1 18 P1 0.203125 1.07015e-05 0.00811408
1 17 N17 0.389375 1 19 P2 0.202188 0.00103718 0.0801131
1 18 P1 0.203125 1 19 P2 0.202188 0.0279074 0.16754
$ plink --bfile mydata --ld P1 P2 --allow-no-sex
R-sq = 0.0279074 D' = 0.16754
Haplotype Frequency Expectation under LE
--------- --------- --------------------
aa 0.068063 0.041069
Aa 0.134124 0.161118
aA 0.135062 0.162056
AA 0.662751 0.635757
In phase alleles are aa/AA
There are other options.
$ plink --bfile mydata --ld rs2840528 rs7545940 # LD between rs2840528 and rx7545940
$ plink --bfile mydata --r2 --ld-window 10 --ld-window-kb 1000 --ld-window-r2 0.2 # these are default values
| - | AA | Aa | aa | O | AA | Aa | aa | O | AA | Aa | aa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BB | 4AB | 2AB, 2aB | 4aB | BB | 726 | 256 | 43 | BB | 0.45375 | 0.16 | 0.026875 |
| Bb | 2AB, 2Ab | AB, Ab, aB, ab | 2aB, ab | Bb | 238 | 245 | 17 | Bb | 0.14875 | 0.153125 | 0.010625 |
| bb | 4Ab | 2Ab, 2ab | 4ab | bb | 49 | 26 | 0 | bb | 0.030625 | 0.01625 | 0 |
| - | A | a | O | A | a | E | A | a |
|---|---|---|---|---|---|---|---|---|
| B | PAB | PaB | B | 0.64640625 | 0.15046875 | B | 0.63575684 | 0.16205566 |
| b | PAb | Pab | b | 0.15140625 | 0.05171875 | b | 0.16111816 | 0.04106934 |
Epistasis
http://zzz.bwh.harvard.edu/plink/epi.shtml
https://www.cog-genomics.org/plink/1.9/epistasis
$ plink --bfile mydata --allow-no-sex --epistasis --epi1 0.0001 --epi2 0.05
# OR_INT (=Odds ratio for interaction),
# STAT (=Chi-square statistic, 1df),
# P (=Asymptotic p-value).
$ less plink.epi.cc
CHR1 SNP1 CHR2 SNP2 OR_INT STAT P
1 P1 1 P2 0.0576579 184.702 4.745e-42
$ less plink.epi.cc.summary
# N_SIG (=Number of significant epistatic tests (p <= "--epi2" threshold)),
# N_TOT (=Number of valid tests (i.e. non-zero allele counts, etc)),
# PROP (=Proportion significant of valid tests).
CHR SNP N_SIG N_TOT PROP BEST_CHISQ BEST_CHR BEST_SNP
1 N0 0 18 0 3.289 1 N9
1 N1 0 19 0 3.486 1 N12
1 N2 0 19 0 3.511 1 N11
1 N3 1 19 0.05263 8.947 1 N7
1 N4 0 19 0 2.694 1 N12
1 N5 2 19 0.1053 7.606 1 N11
1 N6 0 19 0 3.183 1 N0
1 N7 1 19 0.05263 8.947 1 N3
1 N8 1 19 0.05263 11.93 1 N14
1 N9 0 19 0 3.289 1 N0
1 N10 0 18 0 2.215 1 N7
1 N11 1 19 0.05263 7.606 1 N5
1 N12 0 19 0 3.486 1 N1
1 N13 0 19 0 1.812 1 P1
1 N14 1 19 0.05263 11.93 1 N8
1 N15 0 19 0 3.274 1 N0
1 N16 0 19 0 3.507 1 P1
1 N17 0 19 0 2.206 1 N7
1 P1 1 19 0.05263 184.7 1 P2
1 P2 2 19 0.1053 184.7 1 P1
$ plink --bfile mydata --twolocus P1 P2
$ less plink.twolocus
All individuals
===============
P2
a/a a/A A/A 0/0 */*
P1 a/a 0 26 49 0 75
a/A 17 245 238 0 500
A/A 43 256 726 0 1025
0/0 0 0 0 0 0
*/* 60 527 1013 0 1600
P2
a/a a/A A/A 0/0 */*
P1 a/a 0 0.01625 0.030625 0 0.046875
a/A 0.010625 0.153125 0.14875 0 0.3125
A/A 0.026875 0.16 0.45375 0 0.640625
0/0 0 0 0 0 0
*/* 0.0375 0.329375 0.633125 0 1
Cases
=====
P2
a/a a/A A/A 0/0 */*
P1 a/a 0 22 20 0 42
a/A 14 7 232 0 253
A/A 15 235 255 0 505
0/0 0 0 0 0 0
*/* 29 264 507 0 800
P2
a/a a/A A/A 0/0 */*
P1 a/a 0 0.0275 0.025 0 0.0525
a/A 0.0175 0.00875 0.29 0 0.31625
A/A 0.01875 0.29375 0.31875 0 0.63125
0/0 0 0 0 0 0
*/* 0.03625 0.33 0.63375 0 1
Controls
========
P2
a/a a/A A/A 0/0 */*
P1 a/a 0 4 29 0 33
a/A 3 238 6 0 247
A/A 28 21 471 0 520
0/0 0 0 0 0 0
*/* 31 263 506 0 800
P2
a/a a/A A/A 0/0 */*
P1 a/a 0 0.005 0.03625 0 0.04125
a/A 0.00375 0.2975 0.0075 0 0.30875
A/A 0.035 0.02625 0.58875 0 0.65
0/0 0 0 0 0 0
*/* 0.03875 0.32875 0.6325 0 1
Networks
Ternary Plot
Combinations
Max cases
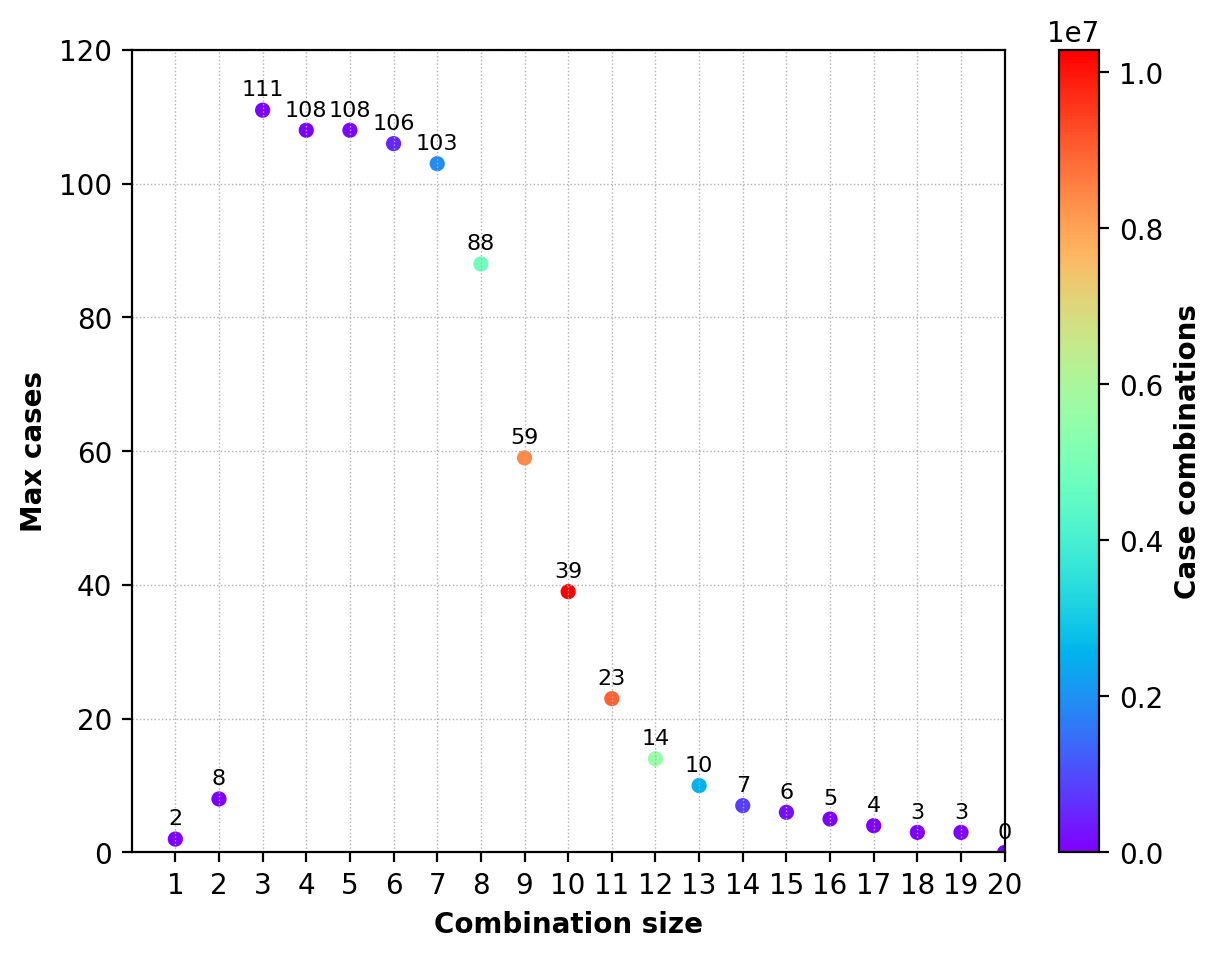
- No_of_SNPs (SNPs) [Genotype] No_of_Sample [Sample_ID]
- 1 ('N13',) [2] 2 ['ncs359' 'ncs725']
- 2 ('N1', 'N5') [2 0] 8 ['ncs30' 'ncs316' 'ncs360' 'ncs426' 'ncs487' 'ncs545' 'ncs593' 'ncs725']
- 3 ('N7', 'P1', 'P2') [1 1 0] 111 "['ncs5' 'ncs6' 'ncs10' 'ncs13' 'ncs22' 'ncs34' 'ncs39' 'ncs44' 'ncs50' 'ncs55' 'ncs62' 'ncs67' 'ncs80' 'ncs83' 'ncs85' 'ncs98' 'ncs100' 'ncs101' 'ncs102' 'ncs106' 'ncs107' 'ncs110' 'ncs111' 'ncs132' 'ncs134' 'ncs137' 'ncs139' 'ncs149' 'ncs154' 'ncs178' 'ncs183' 'ncs185' 'ncs198' 'ncs205' 'ncs209' 'ncs211' 'ncs213' 'ncs216' 'ncs217' 'ncs218' 'ncs226' 'ncs229' 'ncs250' 'ncs266' 'ncs269' 'ncs275' 'ncs276' 'ncs280' 'ncs282' 'ncs291' 'ncs292' 'ncs299' 'ncs307' 'ncs308' 'ncs312' 'ncs319' 'ncs327' 'ncs334' 'ncs341' 'ncs347' 'ncs350' 'ncs365' 'ncs374' 'ncs377' 'ncs383' 'ncs385' 'ncs386' 'ncs388' 'ncs400' 'ncs406' 'ncs412' 'ncs420' 'ncs421' 'ncs427' 'ncs428' 'ncs430' 'ncs432' 'ncs454' 'ncs461' 'ncs471' 'ncs476' 'ncs482' 'ncs487' 'ncs542' 'ncs551' 'ncs552' 'ncs570' 'ncs575' 'ncs584' 'ncs586' 'ncs591' 'ncs595' 'ncs598' 'ncs610' 'ncs635' 'ncs648' 'ncs672' 'ncs673' 'ncs678' 'ncs686' 'ncs690' 'ncs694' 'ncs706' 'ncs716' 'ncs717' 'ncs751' 'ncs754' 'ncs772' 'ncs780' 'ncs785' 'ncs787']"
- 4 ('N0', 'N7', 'P1', 'P2') [0 1 1 0] 108 "['ncs5' 'ncs6' 'ncs10' 'ncs13' 'ncs22' 'ncs34' 'ncs39' 'ncs44' 'ncs50' 'ncs55' 'ncs62' 'ncs67' 'ncs80' 'ncs83' 'ncs85' 'ncs98' 'ncs100' 'ncs101' 'ncs102' 'ncs106' 'ncs107' 'ncs110' 'ncs111' 'ncs132' 'ncs134' 'ncs137' 'ncs139' 'ncs149' 'ncs154' 'ncs178' 'ncs183' 'ncs185' 'ncs198' 'ncs205' 'ncs209' 'ncs211' 'ncs213' 'ncs216' 'ncs217' 'ncs218' 'ncs229' 'ncs250' 'ncs266' 'ncs269' 'ncs275' 'ncs276' 'ncs282' 'ncs291' 'ncs292' 'ncs299' 'ncs307' 'ncs308' 'ncs312' 'ncs319' 'ncs327' 'ncs334' 'ncs341' 'ncs347' 'ncs350' 'ncs365' 'ncs374' 'ncs377' 'ncs383' 'ncs385' 'ncs386' 'ncs388' 'ncs400' 'ncs406' 'ncs412' 'ncs420' 'ncs421' 'ncs427' 'ncs428' 'ncs430' 'ncs432' 'ncs454' 'ncs461' 'ncs471' 'ncs482' 'ncs487' 'ncs542' 'ncs551' 'ncs552' 'ncs570' 'ncs575' 'ncs584' 'ncs586' 'ncs591' 'ncs595' 'ncs598' 'ncs610' 'ncs635' 'ncs648' 'ncs672' 'ncs673' 'ncs678' 'ncs686' 'ncs690' 'ncs694' 'ncs706' 'ncs716' 'ncs717' 'ncs751' 'ncs754' 'ncs772' 'ncs780' 'ncs785' 'ncs787']"
- 5 ('N1', 'N8', 'N10', 'P1', 'P2') [0 0 0 1 0] 108 "['ncs6' 'ncs16' 'ncs24' 'ncs33' 'ncs34' 'ncs39' 'ncs44' 'ncs66' 'ncs67' 'ncs90' 'ncs91' 'ncs94' 'ncs95' 'ncs98' 'ncs100' 'ncs110' 'ncs118' 'ncs132' 'ncs134' 'ncs137' 'ncs154' 'ncs156' 'ncs175' 'ncs185' 'ncs188' 'ncs213' 'ncs216' 'ncs217' 'ncs221' 'ncs226' 'ncs228' 'ncs231' 'ncs232' 'ncs242' 'ncs250' 'ncs256' 'ncs257' 'ncs261' 'ncs266' 'ncs269' 'ncs276' 'ncs281' 'ncs282' 'ncs290' 'ncs291' 'ncs292' 'ncs299' 'ncs300' 'ncs308' 'ncs309' 'ncs319' 'ncs343' 'ncs350' 'ncs365' 'ncs377' 'ncs385' 'ncs387' 'ncs388' 'ncs395' 'ncs400' 'ncs410' 'ncs415' 'ncs420' 'ncs428' 'ncs432' 'ncs433' 'ncs434' 'ncs444' 'ncs451' 'ncs469' 'ncs474' 'ncs476' 'ncs503' 'ncs519' 'ncs528' 'ncs551' 'ncs560' 'ncs567' 'ncs573' 'ncs586' 'ncs600' 'ncs609' 'ncs615' 'ncs617' 'ncs621' 'ncs625' 'ncs635' 'ncs646' 'ncs648' 'ncs673' 'ncs686' 'ncs690' 'ncs706' 'ncs716' 'ncs717' 'ncs721' 'ncs727' 'ncs730' 'ncs734' 'ncs736' 'ncs750' 'ncs778' 'ncs779' 'ncs780' 'ncs785' 'ncs786' 'ncs788' 'ncs795']"
- 6 ('N1', 'N4', 'N10', 'N13', 'P1', 'P2') [0 0 0 0 1 0] 106 "['ncs5' 'ncs16' 'ncs33' 'ncs34' 'ncs39' 'ncs40' 'ncs44' 'ncs46' 'ncs62' 'ncs67' 'ncs90' 'ncs94' 'ncs98' 'ncs100' 'ncs102' 'ncs107' 'ncs118' 'ncs132' 'ncs137' 'ncs154' 'ncs156' 'ncs160' 'ncs167' 'ncs178' 'ncs185' 'ncs188' 'ncs190' 'ncs216' 'ncs217' 'ncs218' 'ncs228' 'ncs229' 'ncs231' 'ncs238' 'ncs242' 'ncs250' 'ncs256' 'ncs257' 'ncs261' 'ncs266' 'ncs275' 'ncs276' 'ncs281' 'ncs282' 'ncs290' 'ncs299' 'ncs307' 'ncs308' 'ncs309' 'ncs319' 'ncs322' 'ncs327' 'ncs343' 'ncs354' 'ncs377' 'ncs378' 'ncs379' 'ncs384' 'ncs386' 'ncs387' 'ncs388' 'ncs395' 'ncs408' 'ncs410' 'ncs415' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs447' 'ncs457' 'ncs469' 'ncs471' 'ncs474' 'ncs476' 'ncs503' 'ncs519' 'ncs528' 'ncs551' 'ncs560' 'ncs573' 'ncs586' 'ncs589' 'ncs591' 'ncs595' 'ncs598' 'ncs600' 'ncs615' 'ncs617' 'ncs641' 'ncs646' 'ncs673' 'ncs678' 'ncs686' 'ncs690' 'ncs711' 'ncs717' 'ncs721' 'ncs727' 'ncs730' 'ncs734' 'ncs754' 'ncs778' 'ncs779' 'ncs786' 'ncs795']"
- 7 ('N0', 'N1', 'N4', 'N10', 'N13', 'P1', 'P2') [0 0 0 0 0 1 0] 103 "['ncs5' 'ncs16' 'ncs33' 'ncs34' 'ncs39' 'ncs40' 'ncs44' 'ncs46' 'ncs62' 'ncs67' 'ncs90' 'ncs94' 'ncs98' 'ncs100' 'ncs102' 'ncs107' 'ncs118' 'ncs132' 'ncs137' 'ncs154' 'ncs156' 'ncs160' 'ncs167' 'ncs178' 'ncs185' 'ncs188' 'ncs190' 'ncs216' 'ncs217' 'ncs218' 'ncs228' 'ncs229' 'ncs231' 'ncs238' 'ncs250' 'ncs256' 'ncs257' 'ncs261' 'ncs266' 'ncs275' 'ncs276' 'ncs281' 'ncs282' 'ncs290' 'ncs299' 'ncs307' 'ncs308' 'ncs309' 'ncs319' 'ncs322' 'ncs327' 'ncs343' 'ncs354' 'ncs377' 'ncs378' 'ncs379' 'ncs384' 'ncs386' 'ncs387' 'ncs388' 'ncs395' 'ncs408' 'ncs410' 'ncs415' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs447' 'ncs457' 'ncs469' 'ncs471' 'ncs474' 'ncs503' 'ncs519' 'ncs528' 'ncs551' 'ncs573' 'ncs586' 'ncs589' 'ncs591' 'ncs595' 'ncs598' 'ncs600' 'ncs615' 'ncs617' 'ncs641' 'ncs646' 'ncs673' 'ncs678' 'ncs686' 'ncs690' 'ncs711' 'ncs717' 'ncs721' 'ncs727' 'ncs730' 'ncs734' 'ncs754' 'ncs778' 'ncs779' 'ncs786' 'ncs795']"
- 8 ('N0', 'N1', 'N4', 'N10', 'N13', 'N14', 'P1', 'P2') [0 0 0 0 0 0 1 0] 88 "['ncs16' 'ncs33' 'ncs34' 'ncs39' 'ncs40' 'ncs44' 'ncs46' 'ncs67' 'ncs90' 'ncs94' 'ncs98' 'ncs102' 'ncs107' 'ncs118' 'ncs132' 'ncs137' 'ncs154' 'ncs156' 'ncs167' 'ncs178' 'ncs185' 'ncs188' 'ncs216' 'ncs217' 'ncs218' 'ncs228' 'ncs229' 'ncs231' 'ncs238' 'ncs250' 'ncs256' 'ncs257' 'ncs261' 'ncs266' 'ncs275' 'ncs276' 'ncs282' 'ncs299' 'ncs307' 'ncs308' 'ncs309' 'ncs322' 'ncs327' 'ncs343' 'ncs354' 'ncs377' 'ncs378' 'ncs379' 'ncs384' 'ncs387' 'ncs388' 'ncs408' 'ncs410' 'ncs415' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs447' 'ncs457' 'ncs469' 'ncs471' 'ncs503' 'ncs519' 'ncs528' 'ncs551' 'ncs573' 'ncs586' 'ncs589' 'ncs591' 'ncs595' 'ncs598' 'ncs600' 'ncs615' 'ncs617' 'ncs641' 'ncs646' 'ncs673' 'ncs678' 'ncs690' 'ncs711' 'ncs717' 'ncs727' 'ncs730' 'ncs754' 'ncs779' 'ncs786' 'ncs795']"
- 9 ('N0', 'N1', 'N4', 'N8', 'N10', 'N13', 'N14', 'P1', 'P2') [0 0 0 0 0 0 0 1 0] 59 "['ncs16' 'ncs33' 'ncs34' 'ncs39' 'ncs44' 'ncs67' 'ncs90' 'ncs94' 'ncs98' 'ncs118' 'ncs132' 'ncs137' 'ncs154' 'ncs156' 'ncs185' 'ncs188' 'ncs216' 'ncs217' 'ncs228' 'ncs231' 'ncs250' 'ncs256' 'ncs257' 'ncs261' 'ncs266' 'ncs276' 'ncs282' 'ncs299' 'ncs308' 'ncs309' 'ncs343' 'ncs377' 'ncs387' 'ncs388' 'ncs410' 'ncs415' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs469' 'ncs503' 'ncs519' 'ncs528' 'ncs551' 'ncs573' 'ncs586' 'ncs600' 'ncs615' 'ncs617' 'ncs646' 'ncs673' 'ncs690' 'ncs717' 'ncs727' 'ncs730' 'ncs779' 'ncs786' 'ncs795']"
- 9 ('N0', 'N1', 'N8', 'N10', 'N13', 'N14', 'N16', 'P1', 'P2') [0 0 0 0 0 0 0 1 0] 59 "['ncs6' 'ncs16' 'ncs24' 'ncs39' 'ncs66' 'ncs67' 'ncs91' 'ncs95' 'ncs98' 'ncs110' 'ncs118' 'ncs132' 'ncs137' 'ncs154' 'ncs185' 'ncs188' 'ncs216' 'ncs221' 'ncs231' 'ncs232' 'ncs256' 'ncs257' 'ncs261' 'ncs269' 'ncs282' 'ncs291' 'ncs299' 'ncs300' 'ncs308' 'ncs343' 'ncs365' 'ncs385' 'ncs387' 'ncs388' 'ncs410' 'ncs415' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs451' 'ncs503' 'ncs528' 'ncs567' 'ncs573' 'ncs586' 'ncs600' 'ncs615' 'ncs617' 'ncs621' 'ncs625' 'ncs635' 'ncs646' 'ncs717' 'ncs727' 'ncs730' 'ncs750' 'ncs785' 'ncs795']"
- 10 ('N0', 'N1', 'N4', 'N8', 'N10', 'N13', 'N14', 'N16', 'P1', 'P2') [0 0 0 0 0 0 0 0 1 0] 39 "['ncs16' 'ncs39' 'ncs67' 'ncs98' 'ncs118' 'ncs132' 'ncs137' 'ncs154' 'ncs185' 'ncs188' 'ncs216' 'ncs231' 'ncs256' 'ncs257' 'ncs261' 'ncs282' 'ncs299' 'ncs308' 'ncs343' 'ncs387' 'ncs388' 'ncs410' 'ncs415' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs503' 'ncs528' 'ncs573' 'ncs586' 'ncs600' 'ncs615' 'ncs617' 'ncs646' 'ncs717' 'ncs727' 'ncs730' 'ncs795']"
- 11 ('N0', 'N1', 'N4', 'N8', 'N10', 'N13', 'N14', 'N16', 'N17', 'P1', 'P2') [0 0 0 0 0 0 0 0 1 1 0] 23 "['ncs16' 'ncs98' 'ncs118' 'ncs132' 'ncs185' 'ncs231' 'ncs257' 'ncs261' 'ncs282' 'ncs299' 'ncs343' 'ncs388' 'ncs415' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs573' 'ncs586' 'ncs600' 'ncs717' 'ncs727' 'ncs730']"
- 12 ('N0', 'N1', 'N3', 'N8', 'N10', 'N12', 'N13', 'N14', 'N15', 'N16', 'P1', 'P2') [0 0 1 0 0 0 0 0 1 0 1 0] 14 "['ncs6' 'ncs24' 'ncs66' 'ncs118' 'ncs132' 'ncs221' 'ncs232' 'ncs282' 'ncs299' 'ncs385' 'ncs410' 'ncs451' 'ncs567' 'ncs795']"
- 12 ('N0', 'N1', 'N4', 'N6', 'N8', 'N10', 'N13', 'N14', 'N16', 'N17', 'P1', 'P2') [0 0 0 0 0 0 0 0 0 1 1 0] 14 "['ncs16' 'ncs231' 'ncs261' 'ncs282' 'ncs299' 'ncs388' 'ncs420' 'ncs428' 'ncs433' 'ncs434' 'ncs573' 'ncs586' 'ncs600' 'ncs727']"
- 13 ('N0', 'N1', 'N2', 'N5', 'N6', 'N8', 'N10', 'N11', 'N12', 'N13', 'N14', 'P1', 'P2') [0 0 1 0 0 0 0 1 0 0 0 1 0] 10 "['ncs24' 'ncs213' 'ncs299' 'ncs377' 'ncs434' 'ncs567' 'ncs573' 'ncs609' 'ncs727' 'ncs780']"
- 13 ('N0', 'N1', 'N4', 'N5', 'N8', 'N10', 'N11', 'N13', 'N14', 'N16', 'N17', 'P1', 'P2') [0 0 0 0 0 0 1 0 0 0 1 1 0] 10 "['ncs98' 'ncs118' 'ncs257' 'ncs299' 'ncs343' 'ncs388' 'ncs415' 'ncs434' 'ncs573' 'ncs727']"
- 14 ('N0', 'N1', 'N2', 'N4', 'N6', 'N7', 'N8', 'N9', 'N10', 'N12', 'N13', 'N14', 'N16', 'P1') [0 0 1 0 0 1 0 0 0 0 0 0 0 0] 7 ['ncs77' 'ncs143' 'ncs176' 'ncs272' 'ncs579' 'ncs623' 'ncs628']
- 14 ('N0', 'N1', 'N2', 'N4', 'N6', 'N8', 'N9', 'N10', 'N12', 'N13', 'N14', 'N15', 'N16', 'P2') [0 0 1 0 0 0 0 0 0 0 0 0 0 0] 7 ['ncs176' 'ncs434' 'ncs573' 'ncs623' 'ncs662' 'ncs684' 'ncs727']
- 14 ('N0', 'N1', 'N2', 'N5', 'N6', 'N8', 'N10', 'N11', 'N12', 'N13', 'N14', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 1 0 0 0 1 1 0] 7 ['ncs213' 'ncs299' 'ncs434' 'ncs567' 'ncs573' 'ncs609' 'ncs727']
- 14 ('N0', 'N1', 'N2', 'N5', 'N6', 'N9', 'N10', 'N11', 'N12', 'N13', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 1 0 0 0 1 1 0] 7 ['ncs5' 'ncs386' 'ncs434' 'ncs567' 'ncs573' 'ncs672' 'ncs727']
- 14 ('N0', 'N3', 'N4', 'N6', 'N8', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N17', 'P1', 'P2') [0 1 0 1 0 0 1 0 0 0 1 1 1 0] 7 ['ncs34' 'ncs118' 'ncs426' 'ncs610' 'ncs724' 'ncs751' 'ncs787']
- 15 ('N0', 'N1', 'N2', 'N4', 'N6', 'N7', 'N8', 'N9', 'N10', 'N12', 'N13', 'N14', 'N16', 'N17', 'P2') [0 0 1 0 0 1 0 0 0 0 0 0 0 1 0] 6 ['ncs143' 'ncs176' 'ncs272' 'ncs428' 'ncs579' 'ncs623']
- 15 ('N0', 'N1', 'N2', 'N4', 'N6', 'N8', 'N9', 'N10', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P2') [0 0 1 0 0 0 0 0 0 0 0 0 0 1 0] 6 ['ncs176' 'ncs434' 'ncs573' 'ncs623' 'ncs662' 'ncs727']
- 16 ('N0', 'N1', 'N2', 'N4', 'N6', 'N7', 'N8', 'N9', 'N10', 'N12', 'N13', 'N14', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 1 0 0 0 0 0 0 0 1 0 0] 5 ['ncs143' 'ncs176' 'ncs272' 'ncs579' 'ncs623']
- 16 ('N0', 'N1', 'N2', 'N4', 'N6', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P2') [0 0 1 0 0 0 0 0 1 0 0 0 0 0 1 0] 5 ['ncs176' 'ncs434' 'ncs573' 'ncs662' 'ncs727']
- 17 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 0] 4 ['ncs176' 'ncs434' 'ncs573' 'ncs727']
- 17 ('N0', 'N1', 'N2', 'N4', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 0] 4 ['ncs434' 'ncs573' 'ncs662' 'ncs727']
- 17 ('N0', 'N1', 'N2', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 1 1 0] 4 ['ncs434' 'ncs567' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N3', 'N4', 'N5', 'N6', 'N7', 'N8', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N17', 'P1', 'P2') [0 1 1 1 0 0 1 1 0 0 1 0 0 0 1 1 1 0] 3 ['ncs610' 'ncs751' 'ncs787']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N5', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N4', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N1', 'N2', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727'] -18 ('N0', 'N1', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N0', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 18 ('N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']
- 19 ('N0', 'N1', 'N2', 'N4', 'N5', 'N6', 'N7', 'N8', 'N9', 'N10', 'N11', 'N12', 'N13', 'N14', 'N15', 'N16', 'N17', 'P1', 'P2') [0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0] 3 ['ncs434' 'ncs573' 'ncs727']